Examples
1CZY
LMP1 binding peptide and the TNFR-associated factor 2
In this example of protein-peptide docking, we select in step 2 a couple of residue restraints in the receptor partner (TNRF2) (A.411 and A.466).
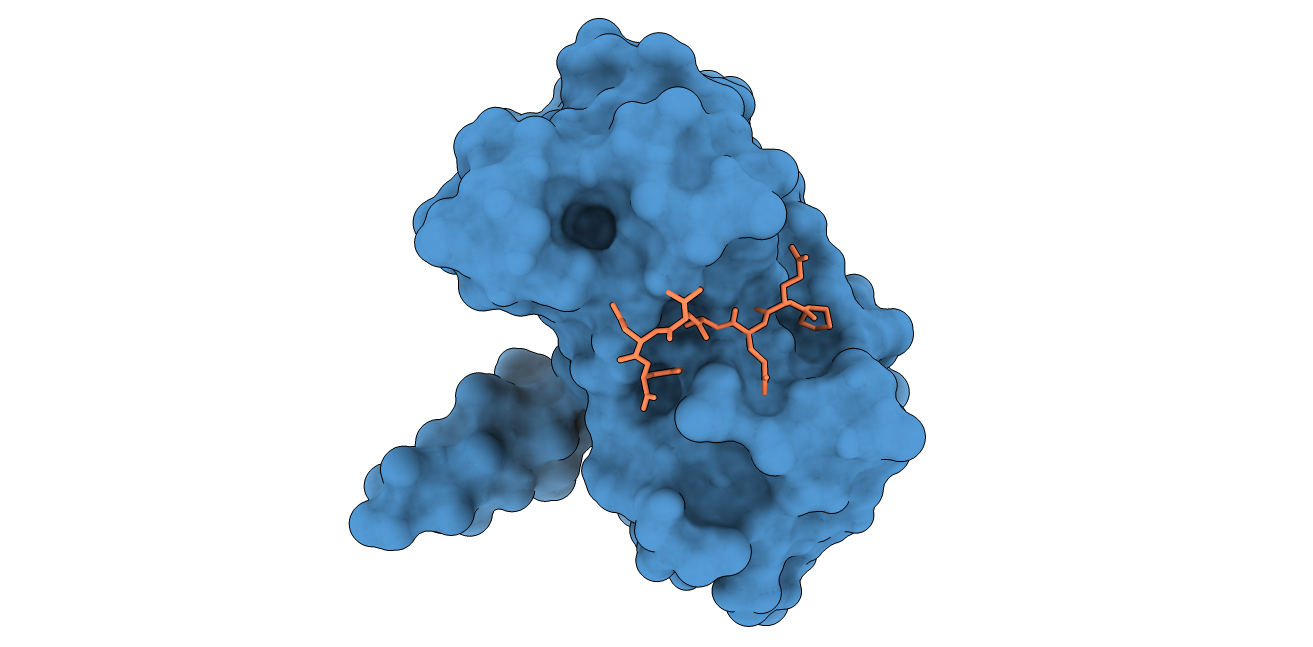 In step 3, we enable flexibility on both receptor and ligand partners.
In step 3, we enable flexibility on both receptor and ligand partners.
1J1V
DnaA domainIV complexed with DnaAbox DNA
In this example of protein-DNA docking, we select in step 2 a single residue restraint to drive the docking in the receptor partner (DnaA domainIV) (A.443).
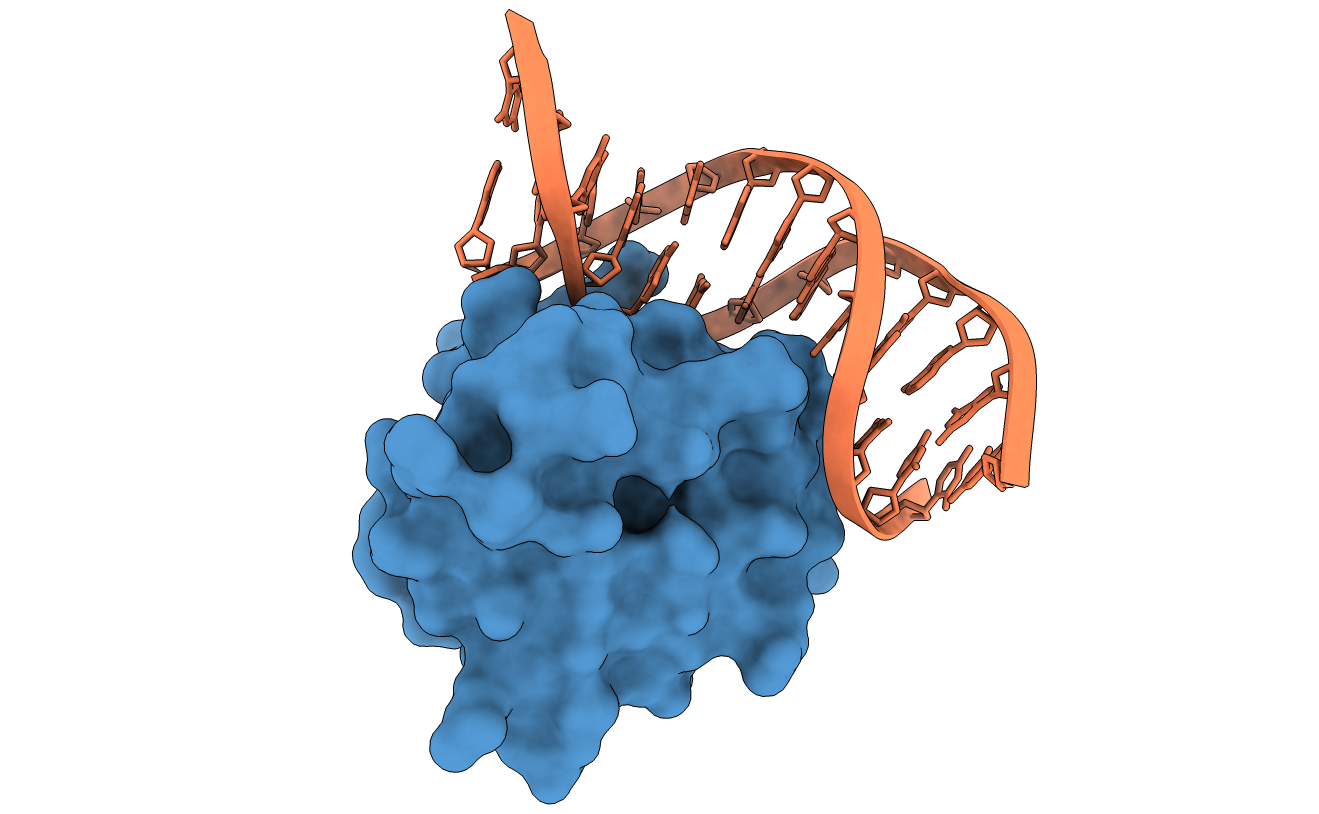 In step 3, we enable flexibility on both receptor and ligand partners.
In step 3, we enable flexibility on both receptor and ligand partners.
3MXW
Inhibition of the Sonic Hedgehog metalloprotease by the anti-Shh 5E1 antibody
In this example of antibody-antigen docking, we show the automatic antibody mode to drive the docking. We select as receptor partner the anti-Shh 5E1 antibody molecule and the Sonic Hedgehog metalloprease as the ligand partner. After selecting the correct numbering of the input antibody PDB file (Chothia), in step 3 residue restraints are displayed in pink color, corresponding with the CDR loops.
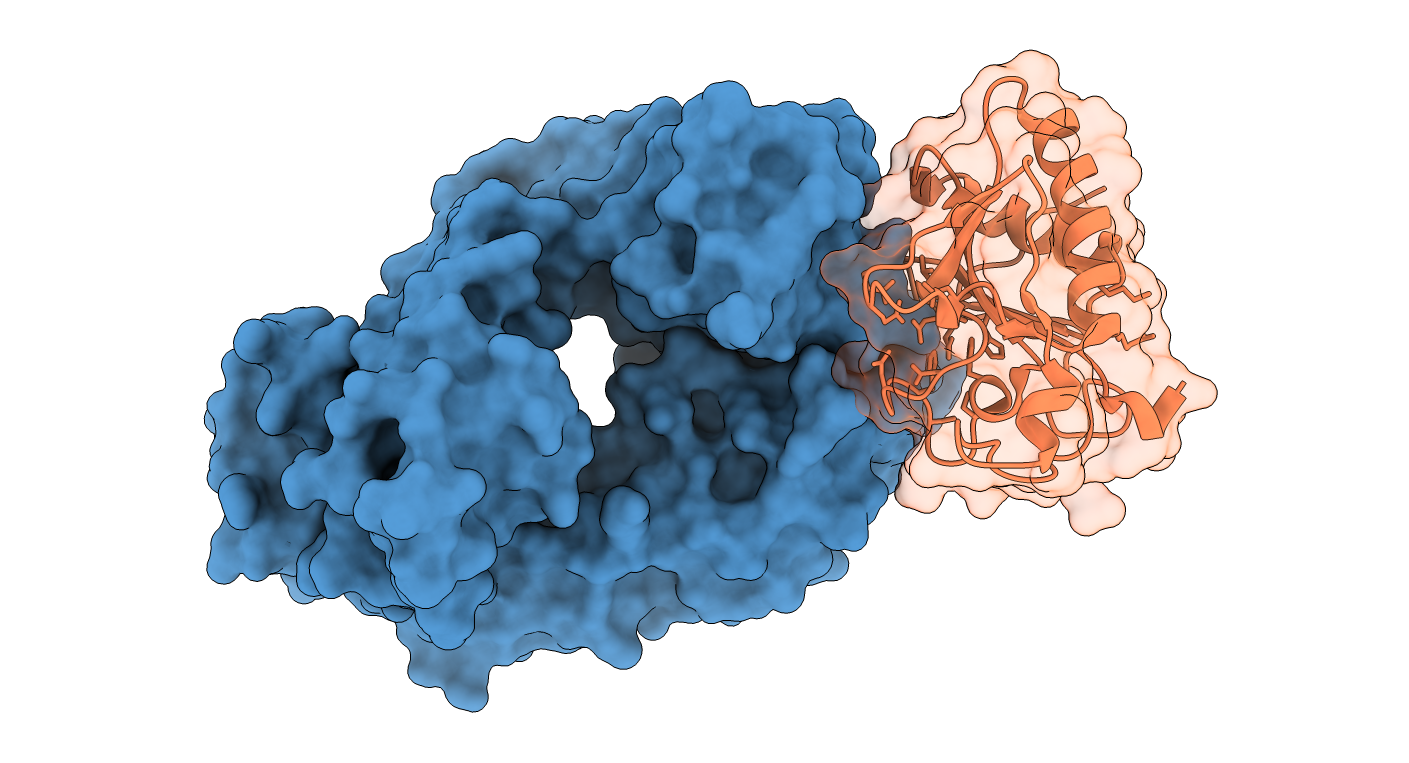 Flexibility for both partners is activated in step 3.
Flexibility for both partners is activated in step 3.
3X29
Integrative modeling of Claudin-19 in complex with the C-CPE enterotoxin
In this integrative modeling example, we make use of a simulated coarse-grained bi-lipidic membrane as explained in the official LightDock tutorial and published in Integrative Modeling of Membrane-associated Protein Assemblies (Roel-Touris J, Jiménez-García B. and Bonvin AMJJ.) Nat Commun 11, 6210 (2020).
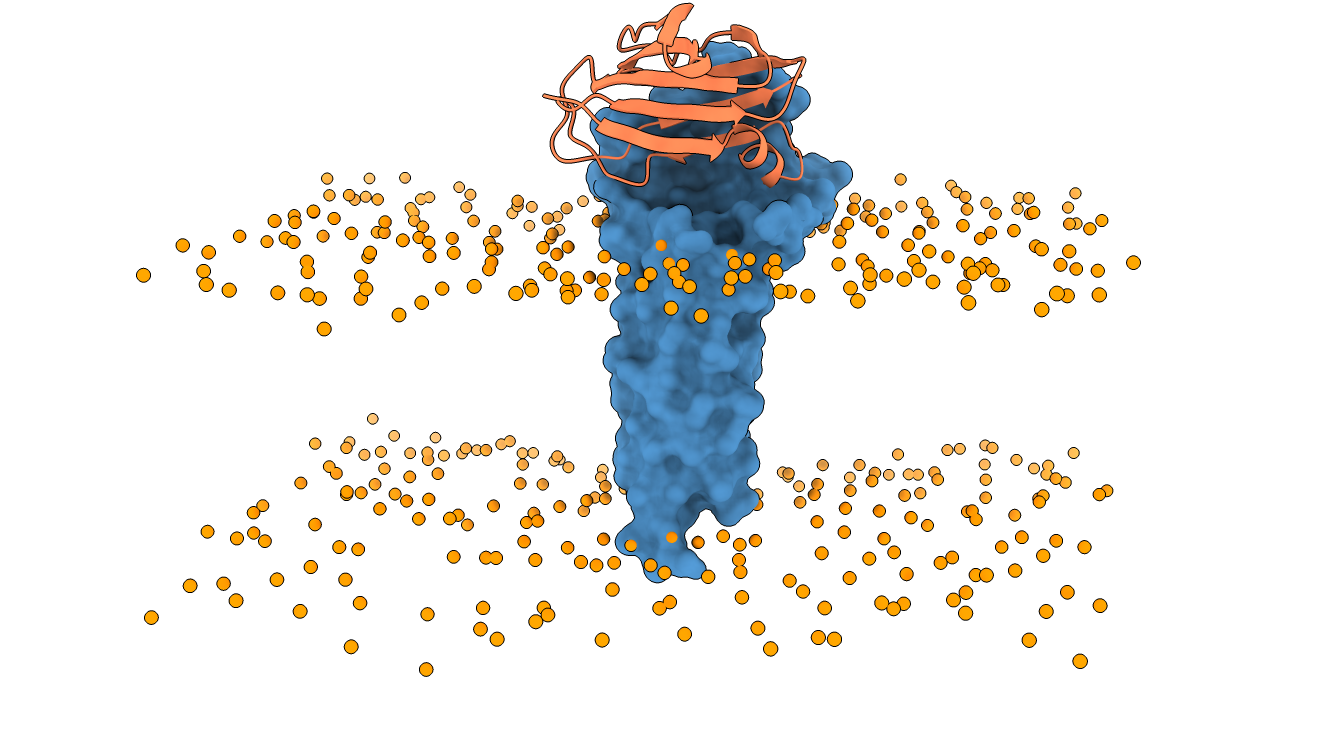 Once the provided receptor PDB file is uploaded to the server, we select the molecule type as
Once the provided receptor PDB file is uploaded to the server, we select the molecule type as membrane. No further restraints are selected in this example.